# A tibble: 24,630 × 5
lsoa_code lsoa_name gen_health_code gen_health_cat observation
<chr> <chr> <dbl> <chr> <dbl>
1 E01011949 Hartlepool 009A 1 Very good health 766
2 E01011949 Hartlepool 009A 2 Good health 567
3 E01011949 Hartlepool 009A 3 Fair health 310
4 E01011949 Hartlepool 009A 4 Bad health 150
5 E01011949 Hartlepool 009A 5 Very bad health 30
6 E01011950 Hartlepool 008A 1 Very good health 368
7 E01011950 Hartlepool 008A 2 Good health 364
8 E01011950 Hartlepool 008A 3 Fair health 185
9 E01011950 Hartlepool 008A 4 Bad health 96
10 E01011950 Hartlepool 008A 5 Very bad health 21
# ℹ 24,620 more rowsExercise 1a: Mapping General Health Categories across Local Authority Districts
Parallelising R workflows
Dataset background
In this exercise, we’ll be working with a subset of Office of National Statistics (ONS) 2021 census data, specifically the responses to the census question on general health. The General Health data was accessed through the ONS data portal in April 2023 and is available through a Open Government Licence v3.0.
The definition of the question according to the ONS is as follows:
General Health
A person’s assessment of the general state of their health from very good to very bad. This assessment is not based on a person’s health over any specified period of time.
As described, the assessment is categorical and each respondent responds with a category from one of the following:
- Very good health
- Good health
- Fair health
- Bad health
- Very bad health
The data we’ll be working with are reported at the Lower layer Super Output Areas (LSOA). The definition of an LSOA is:
Lower layer Super Output Areas (4,926 of 35,672)
Lower layer Super Output Areas (LSOAs) comprise between 400 and 1,200 households and have a usually resident population between 1,000 and 3,000 persons. We will be working with a subset of 4,926 LSOAs.
Workflow objective
The objective of the workflow is to produce maps to visualise the distribution and other metrics of General Health categories across LSOA respondents at the lower tier Local District Authority (LAD) level.
Lower tier Local District Authorities (40 of 331)
Lower tier local authorities (LADs) provide a range of local services. There are 309 lower tier local authorities in England made up of 181 non-metropolitan districts, 59 unitary authorities, 36 metropolitan districts and 33 London boroughs (including City of London). In Wales there are 22 local authorities made up of 22 unitary authorities. We will be working with a subset of 40 LADs.
Exercise materials
The materials for this exercise are contained in the health_data/ directory in the course materials.
The data used in this exercise are contained in the data/ subfolder.
Source: Office for National Statistics
License: Open Government Licence v.3.0
Description:
This dataset provides Census 2021 estimates that classify usual residents in England and Wales by the state of their general health. The estimates are as at Census Day, 21 March 2021.
# A tibble: 4,926 × 4
lsoa_code lsoa_name lad_code lad_name
<chr> <chr> <chr> <chr>
1 E01011949 Hartlepool 009A E06000001 Hartlepool
2 E01011950 Hartlepool 008A E06000001 Hartlepool
3 E01011951 Hartlepool 007A E06000001 Hartlepool
4 E01011952 Hartlepool 002A E06000001 Hartlepool
5 E01011953 Hartlepool 002B E06000001 Hartlepool
6 E01011954 Hartlepool 001A E06000001 Hartlepool
7 E01011955 Hartlepool 003A E06000001 Hartlepool
8 E01011957 Hartlepool 003C E06000001 Hartlepool
9 E01011959 Hartlepool 014A E06000001 Hartlepool
10 E01011960 Hartlepool 014B E06000001 Hartlepool
# ℹ 4,916 more rowsSource: Office for National Statistics
License: Open Government Licence v.3.0
Description:
A lookup between Lower layer Super Output Areas (LSOA) and Local Authority Districts (LAD) as at 31 December 2021 in England and Wales.
Simple feature collection with 10 features and 1 field
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: 531936.7 ymin: 180733.9 xmax: 545296.2 ymax: 184928
Projected CRS: OSGB36 / British National Grid
# A tibble: 10 × 2
lsoa_code geometry
<chr> <MULTIPOLYGON [m]>
1 E01000001 (((532151.5 181867.4, 532152.5 181864.5, 532162.5 181867.8, 532175…
2 E01000002 (((532634.5 181926, 532632 181913.4, 532619.1 181847.2, 532618.7 1…
3 E01000003 (((532153.7 182165.2, 532158.2 182151.1, 532158.8 182151.3, 532160…
4 E01000005 (((533619.1 181402.4, 533639.9 181380.2, 533646.6 181373, 533659.4…
5 E01000006 (((545126.9 184310.8, 545145.2 184295.2, 545145.5 184295, 545147.1…
6 E01000007 (((544173 184701.4, 544180.2 184700.2, 544180.3 184700.6, 544185.6…
7 E01000008 (((543719.9 184612.6, 543748.2 184590.3, 543763.8 184578, 543762.5…
8 E01000009 (((544602.7 184628.2, 544605.2 184626.5, 544606.6 184625.3, 544608…
9 E01000011 (((544608 184727.8, 544620.7 184717.8, 544631.5 184709.3, 544639.6…
10 E01000012 (((544113.7 184901.6, 544129.7 184882.7, 544139.4 184888.1, 544146…Source: Office for National Statistics
License: Open Government Licence v.3.0. Contains OS data © Crown copyright and database right [2023]
Description:
This file contains the digital vector boundaries for Lower layer Super Output Areas in England and Wales as at 21 March 2021. The boundaries available are: Full Clipped - clipped to the coastline (Mean High Water (MHW)) mark.
Exercise Workflow
The workflow is based on a typical tidyverse workflow that uses package purrr and is made up of two scripts:
This script runs the main workflow. The steps involved include:
- Load libraries.
- Source custom functions.
- Prepare output directory.
- Load data.
- Join data into a single
sfobject. - Perform some data munging.
- Create index to subset the data to map.
- Split the data into a list of
sfs containing data for an individual LAD and subsets. - Walk the plotting function using
purrracross each element of the split data and save a health data map for each LAD to.png.
# Load Libraries ----
library(sf)
library(dplyr)
library(assertr)
library(ggplot2)
library(colorspace)
library(ggpubr)
# Source function ----
source(here::here("health_data", "R", "map-function.R"))
# Prepare output directory ----
out_dir <- here::here("health_data", "outputs", "maps")
fs::dir_create(out_dir)
# Load data ----
health_data <- readr::read_csv(
here::here(
"health_data", "data",
"lsoa-general_health.csv"
)
)
look_up <- readr::read_csv(
here::here(
"health_data", "data",
"output_area_lookup.csv"
)
)
boundaries <- read_sf(
here::here(
"health_data", "data",
"lsoa_boundaries.geojson"
)
)
# Merge data and validate ----
all_data <- left_join(health_data, look_up,
relationship = "many-to-one"
) %>%
left_join(boundaries, relationship = "many-to-one") %>%
st_as_sf() %>%
assert(not_na, lsoa_code, lsoa_name, lad_name, lad_code, geometry) %>%
verify(inherits(., "sf"))
# Process data ----
# Get ordered health category levels
health_cat_levels <- health_data %>%
select(gen_health_code, gen_health_cat) %>%
distinct() %>%
arrange(gen_health_code) %>%
pull(gen_health_cat)
# Create addition variables obs_perc & z_score, cast gen_health_cat to factor
all_data <- all_data %>%
mutate(gen_health_cat = factor(gen_health_cat,
levels = health_cat_levels
)) %>%
group_by(lsoa_code) %>%
mutate(obs_perc = observation / sum(observation) * 100) %>%
ungroup() %>%
group_by(gen_health_cat) %>%
mutate(z_score = (obs_perc - mean(obs_perc)) / sd(obs_perc)) %>%
ungroup()
# Create iteration indexes ----
idx_start <- 1
idx_end <- 5
idx <- idx_start:idx_end
# Split and subset data ----
split_data <- split(all_data, f = all_data$lad_code)[idx]
# Create maps ----
# Iterate plotting function over each LAD plot data chunk in idx
tictoc::tic()
purrr::walk(
split_data,
~ plot_lad_map(.x, out_dir)
)
cli::cli_h1("Job Complete")
cli::cli_alert_success("{length(idx)} map{?s} written to {.path {out_dir}}.")
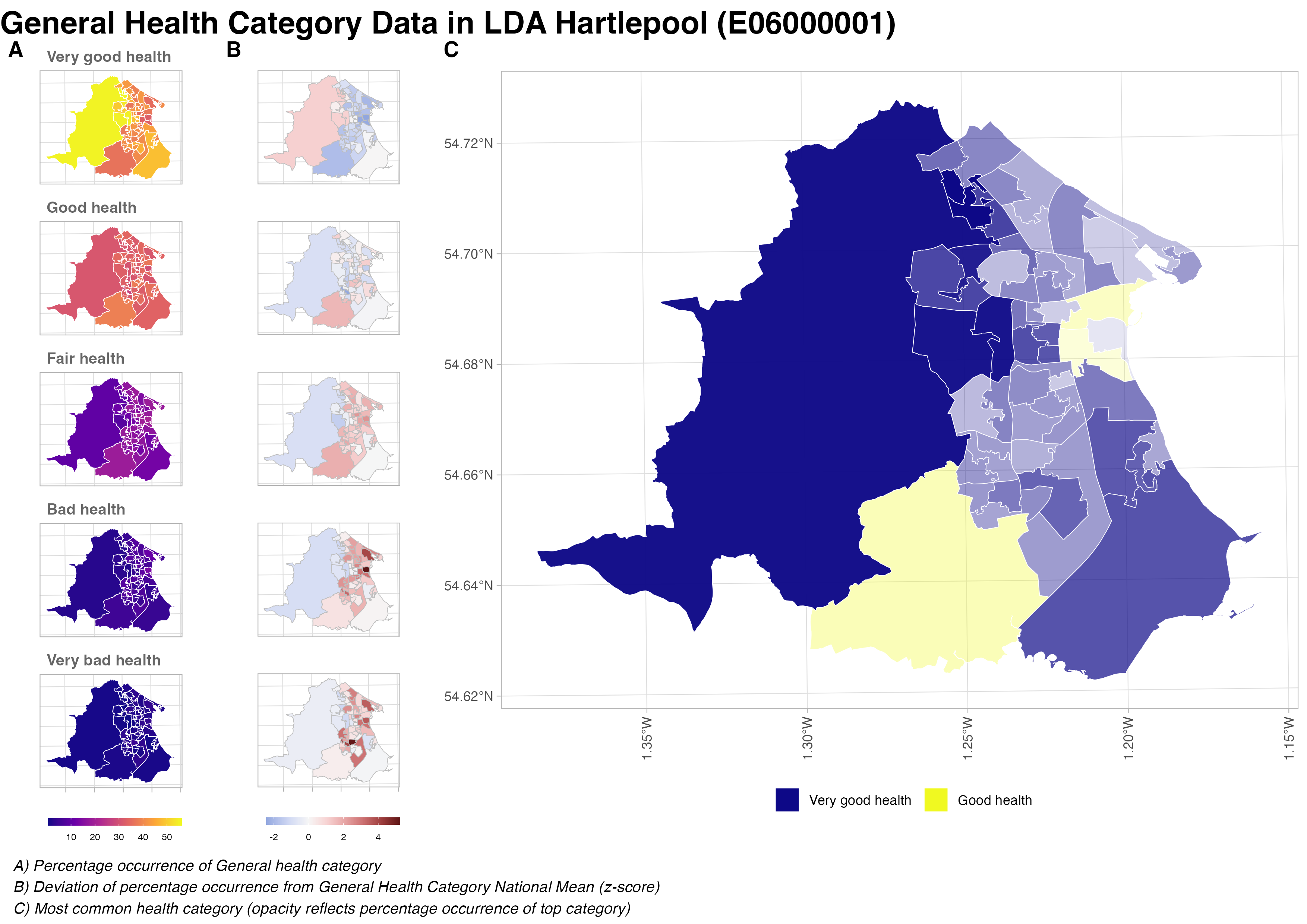
cli::cli_h2("Total time elapsed: {.val {tictoc::toc(quiet = TRUE)$callback_msg}}")This file contains the main mapping function.
It creates 3 maps,
- A facet grid of LSOA level percentage occurrence of each general health category.
- A facet grid of LSOA level deviation of percentage occurrence from each general health category’s national mean (z-score).
- A main map of the most common health category (with the opacity reflecting percentage occurrence of top category).
#' Plot General Health Category maps
#'
#' Plots multiple maps of metrics associated with LSOA level ONS General Health
#' Category responses at the level of an individual Local Authority District.
#' @param plot_data data.frame containing LSOA level plot data subsets for a individual Local Authority.
#' @param out_dir path to the output directory where PNGs of maps will be written to.
#'
#' @return the function is used primarily for it's side effects of plotting a
#' number of plots, combining them and saving as an A4 png to the directory specified
#' by `output_dir`. The output file name for each PNG matches the pattern:
#' `<LSOA_code>_<LSOA_name>_<date_created>.png`
plot_lad_map <- function(plot_data, out_dir) {
tictoc::tic()
lad_code <- unique(plot_data$lad_code)
lad_name <- unique(plot_data$lad_name)
pid <- Sys.getpid()
node <- system2("hostname", stdout = TRUE)
# Create facet plot maps of percentage occurrence of each category across each LSOA.
plt_1 <- plot_data %>%
ggplot() +
geom_sf(aes(fill = obs_perc), colour = "white") +
facet_wrap(~gen_health_cat, ncol = 1) +
scale_fill_viridis_c(name = "", option = "C") +
theme_light() +
theme(
axis.text.x = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
legend.key.height = unit(5, units = "native"),
legend.position = "bottom",
legend.text = element_text(size = 6),
legend.title = element_text(size = 10),
strip.background = element_rect(fill = "white", colour = "white"),
strip.text.x = element_text(colour = "gray40", face = "bold", size = 10, hjust = 0)
)
# Create deviation from national mean percentage occurrence (z-score) facet plot maps
# of each category across each LSOA.
plt_2 <- plot_data %>%
ggplot() +
geom_sf(aes(fill = z_score), colour = "gray") +
scale_fill_continuous_diverging(
palette = "Blue-Red 3",
name = ""
) +
facet_wrap(~gen_health_cat, ncol = 1) +
theme_light() +
theme(
axis.text.x = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
legend.position = "bottom",
legend.text = element_text(size = 6),
legend.key.height = unit(5, units = "native"),
strip.background = element_rect(fill = "white", colour = "white"),
strip.text.x = element_text(colour = "white", face = "bold", size = 10, hjust = 0)
)
# Create map of category of maximum occurrence across each LSOA and scale opacity
# according to maximum category occurence.
plot_main <- plot_data %>%
group_by(lsoa_code) %>%
summarise(
max_cat = head(gen_health_cat[obs_perc == max(obs_perc)], 1),
obs_max_perc = max(obs_perc)
) %>%
ggplot() +
geom_sf(aes(fill = max_cat, alpha = obs_max_perc), colour = "white") +
scale_fill_viridis_d(name = "", option = "C") +
theme_light() +
theme(
axis.text.x = element_text(angle = 90, vjust = 0.5, hjust = 1),
legend.position = "bottom"
) +
guides(alpha = "none")
# Arrange plots
plot <- ggarrange(plt_1, plt_2, plot_main,
labels = c("A", "B", "C"),
ncol = 3, nrow = 1, widths = c(1, 1, 4)
)
# Add plot annotation
plot <- annotate_figure(plot,
top = text_grob(glue::glue("General Health Category Data in LDA {unique(plot_data$lad_name)} ({unique(plot_data$lad_code)})"),
face = "bold",
size = 20,
just = c("left", "bottom"), x = 0, y = 0
),
bottom = text_grob(
paste("A) Percentage occurrence of General health category",
"B) Deviation of percentage occurrence from General Health Category National Mean (z-score)",
"C) Most common health category (opacity reflects percentage occurrence of top category)",
sep = "\n"
),
just = c("left", "top"), x = 0.01, y = 0.99, face = "italic", size = 10, lineheight = 1.15
)
)
file_name <- glue::glue("{lad_code}_{janitor::make_clean_names(lad_name)}_{Sys.Date()}.png")
out_path <- fs::path(file_name, ext = "png")
# Save combined plot
ggsave(
filename = file_name,
plot = plot,
device = "png",
path = out_dir,
width = 29.7, height = 21, units = "cm",
bg = "white"
)
cli::cli_alert_success(c(
"Map for LAD {.val {lad_code} {lad_name}} completed ",
"successfuly on PID {.val {pid}} on {node} ",
"({tictoc::toc(quiet = TRUE)$callback_msg})"
))
}Here’s an example of map output:
Run through workflow sequentially
Let’s open generate-maps-purrr.R and run through the workflow to familiarise ourselves with what it does.
The script is pre set through variables idx_start and idx_end to plot five maps for the first five LADs.
Sourcing the file should create an outputs/maps directory in the health_data directory and save the five maps into it.
The output also gives us information about the time it takes to create each map as well as the total mapping workflow.
The output also makes clear that the workflow is running sequentially, both because the the PID is the same for each map as well as the total time elapsed being the sum of the elapsed time for each individual map.
✔ Map for LAD "E06000001 Hartlepool" completed successfuly on PID 39305 on dcs33297 (1.937 sec elapsed)
✔ Map for LAD "E06000002 Middlesbrough" completed successfuly on PID 39305 on dcs33297 (2.407 sec elapsed)
✔ Map for LAD "E06000003 Redcar and Cleveland" completed successfuly on PID 39305 on dcs33297 (2.497 sec elapsed)
✔ Map for LAD "E06000004 Stockton-on-Tees" completed successfuly on PID 39305 on dcs33297 (3.063 sec elapsed)
✔ Map for LAD "E06000005 Darlington" completed successfuly on PID 39305 on dcs33297 (2.176 sec elapsed)
── Job Complete ─────────────────────────────────
✔ 5 maps written to /Users/Anna/r-rse-parallel-r-materials-02dc656/health_data/outputs/maps.
── Total time elapsed: "12.235 sec elapsed" ──Parallelise workflow using furrr
Let’s now make our mapping workflow able to run in parallel by using package furrr.
Copy generate-maps-purrr.R
Let’s make a copy of generate-maps-purrr.R for us to edit in the same health_data/ directory and name it generate-maps-furrr.R
Next, let’s start editing generate-maps-furrr.R.
Make sure you are editing generate-maps-furrr.R and not generate-maps-purrr.R. To be sure it might be easiest to close generate-maps-purrr.R.
Load the furrr 📦
To start, let’s add a library() call to load the furrr library at the top of the script where libraries are loaded.
Use future_walk instead of walk
The workflow section amenable to parallelisation is the mapping section which iterates over each LAD data chunk.
Each iteration of the plot_lad_map() function takes a small subset of the data and produces a single output (a png) which is completely independent from what’s going on in any of the other iterations. This is a classic example of embarrassingly data parallel problems.
To make our workflow parallelisable, we therefore focus on the purrr::walk() function. As we’ve discussed, the equivalent 📦 in the futureverse to the purrr 📦 is the furrr 📦.
So let’s switch out the purrr::walk() function for the furrr::future_walk() function.
plan(sequential)
future_walk(
split_data,
~ plot_lad_map(.x, out_dir),
.progress = TRUE,
.options = furrr_options(seed = TRUE)
)The function arguments are the same apart from additional future_walk arguments .progress = TRUE which will print a progress bar and .options = furrr_options(seed = TRUE) which specifies the type of seed to be used. For more details, have a look at the future_walk() documentation.
Use sequential plan
Note that we also add a call to the plan function before calling future_walk().
We start by specifying a sequential plan with plan(sequential) to compare to the purrr::walk version.
Let’s re-run the workflow (you can skip the initial data munging and splitting if you want as those steps have not changed).
✔ Map for LAD "E06000001 Hartlepool" completed successfuly on PID 39305 on dcs33297 (1.868 sec elapsed)
✔ Map for LAD "E06000002 Middlesbrough" completed successfuly on PID 39305 on dcs33297 (2.402 sec elapsed)
✔ Map for LAD "E06000003 Redcar and Cleveland" completed successfuly on PID 39305 on dcs33297 (2.545 sec elapsed)
✔ Map for LAD "E06000004 Stockton-on-Tees" completed successfuly on PID 39305 on dcs33297 (3.02 sec elapsed)
✔ Map for LAD "E06000005 Darlington" completed successfuly on PID 39305 on dcs33297 (2.159 sec elapsed)
── Job Complete ─────────────────────────────────
✔ 5 maps written to /Users/Anna/r-rse-parallel-r-materials-02dc656/health_data/outputs/maps.
── Total time elapsed: "12.274 sec elapsed" ──The output confirms that although we have switched to future_walk(), the workflow is functionally the same because we’re using a sequential plan.
Use multisession plan
Now let’s switch to using a parallel plan by substituting plan(sequential) with:
plan(multisession)Progress: ────────────────────────────────────────────────────────────────── 100%
✔ Map for LAD "E06000001 Hartlepool" completed successfuly on PID 79882 on dcs33297 (2.798 sec elapsed)
✔ Map for LAD "E06000002 Middlesbrough" completed successfuly on PID 79881 on dcs33297 (3.028 sec elapsed)
✔ Map for LAD "E06000003 Redcar and Cleveland" completed successfuly on PID 79880 on dcs33297 (3.072 sec elapsed)
✔ Map for LAD "E06000004 Stockton-on-Tees" completed successfuly on PID 79877 on dcs33297 (3.583 sec elapsed)
✔ Map for LAD "E06000005 Darlington" completed successfuly on PID 79875 on dcs33297 (2.658 sec elapsed)
── Job Complete ───────────────────────────────────────────────────────────────
✔ 5 maps written to /Users/Anna/r-rse-parallel-r-materials-02dc656/health_data/outputs/maps.
── Total time elapsed: "6.456 sec elapsed" ──As we can see, our code is now running in parallel as indicated by the fact that the PID used for each map is different. We also managed to make the mapping section of our workflow almost twice faster.
So, we’ve managed to make a small edit to our code and enable it to switch from sequential to parallel with a simple change in the plan.
We’ve successfully managed to:
Examine a classic example of an embarrassingly parallel problem.
Use the
furrr📦 to paralellisepurrr::walk()workflows.